In this tutorial, we will show you how to build a random forest model in SAS.
Random Forest is a machine learning algorithm used for both classification and regression tasks. It combines multiple decision trees to create a stronger, more robust model. Random Forest is particularly effective because it reduces the risk of overfitting and improves generalization compared to single decision trees.
Steps to Build a Random Forest in SAS
Here we are using the built-in SAS dataset named sashelp.heart that contains information related to heart disease. The data set contains 5,209 observations. The variable "status" refers to status of the person whether the person is alive or dead.
Split Data into Training and Test Data
First we split the data into training and test datasets. 70% of data goes to training data, while the other 30% data goes to test dataset. We are using the PROC SURVEYSELECT procedure which is used to perform stratified random sampling on the sorted dataset heart. The stratified sampling ensures that the distribution of the dependent variable remains the same in both training and test datasets.
proc sort data= sashelp.heart out=heart; by status; run; proc surveyselect data=heart rate=0.7 outall out=heart2 seed=1234; strata status; run; data heart_train heart_test; set heart2; if selected = 1 then output heart_train; else output heart_test; drop selected; run; proc freq data=heart_train; table status; run; proc freq data=heart_test; table status; run;
Model Development
In SAS, the HPFOREST procedure is a high-performance procedure to create a random forest model.
PROC HPFOREST DATA=heart_train maxtrees=500 vars_to_try=3 seed=1234 trainfraction=0.7;
target status/ level=binary;
input Weight Height AgeAtStart Smoking Sex / level = binary;
SCORE OUT=scored;
SAVE FILE='/home/deepanshu88us0/heart_forest.bin';
ods output FitStatistics = fitstats;
ods output VariableImportance = Variable_Importance;
run;
The LEVEL option in the TARGET statement defines the dependent variable as binary, nominal, or interval. Whereas, the LEVEL option for the INPUT statement defines the independent variables as binary, nominal, ordinal, or interval.
MAXTREES: Defines the maximum number of trees for a random forest.TRAINFRACTION: Specifies the percentage of training data to train each tree.SEED: Sets the randomization seed for bootstrapping and feature selection.VARS_TO_TRY: Specifies the randomized number of inputs to select at each node.LEAFSIZE: Indicates the minimum number of observations allowed in each branch.ALPHA: Specifies the p-value threshold a candidate variable must meet for a node to be split.MAXDEPTH: Specifies the number of splitting rules for the nodes.PRESELECT: Indicates the method of selecting a splitting feature.
Make sure you specify your location in the FILE= option in SAVE statement. It will be used for applying the random forest to test dataset.
Random Forest algorithm also returns variable importance table. In the above code, it is saved in the dataset named "Variable_Importance". Variable Importance measures the contribution of individual input variables in a model towards making accurate predictions. It helps to understand which variables have the most influence on the model's predictions.
Generating Predictions
The HP4SCORE procedure scores a data set using a random forest model that was previously trained by the HPFOREST procedure. In simple words, it applies the model to the test dataset. It creates the predicted probability column. In this case, it is P_StatusAlive column in the "heart_test_scored" data set.
proc hp4score data=heart_test; id status; score file='/home/deepanshu88us0/heart_forest.bin' out=heart_test_scored; run;
Model Evaluation
After a machine learning model has been trained on a training dataset and possibly fine-tuned using a validation dataset, it's important to assess how well it performs on new, unseen data (test data).
The following SAS code calculates the Area Under Curve (AUC) and Gini coefficient based on the output from a Wilcoxon test.
%let dependent_var = status; %let score_dataset = heart_test_scored; %let score_column = P_StatusAlive; ods output WilcoxonScores=WilcoxonScore; proc npar1way wilcoxon data= &score_dataset.; class &dependent_var.; var &score_column.; run; data AUC; set WilcoxonScore end=eof; retain v1 v2 1; if _n_=1 then v1=abs(ExpectedSum - SumOfScores); v2=N*v2; if eof then do; d=v1/v2; Gini=d * 2; AUC=d+0.5; put AUC= GINI=; keep AUC Gini; output; end; proc print noobs; run;
Result: AUC=0.7564055164 Gini=0.5128110327
Fine Tune Random Forest Model in SAS
In the Random Forest algorithm, the number of variables are randomly selected at each node when building a decision tree within the Random Forest ensemble. It is a hyperparameter that needs to be tuned during the model selection process. In SAS, it is the VARS_TO_TRY= option in PROC HPFOREST.
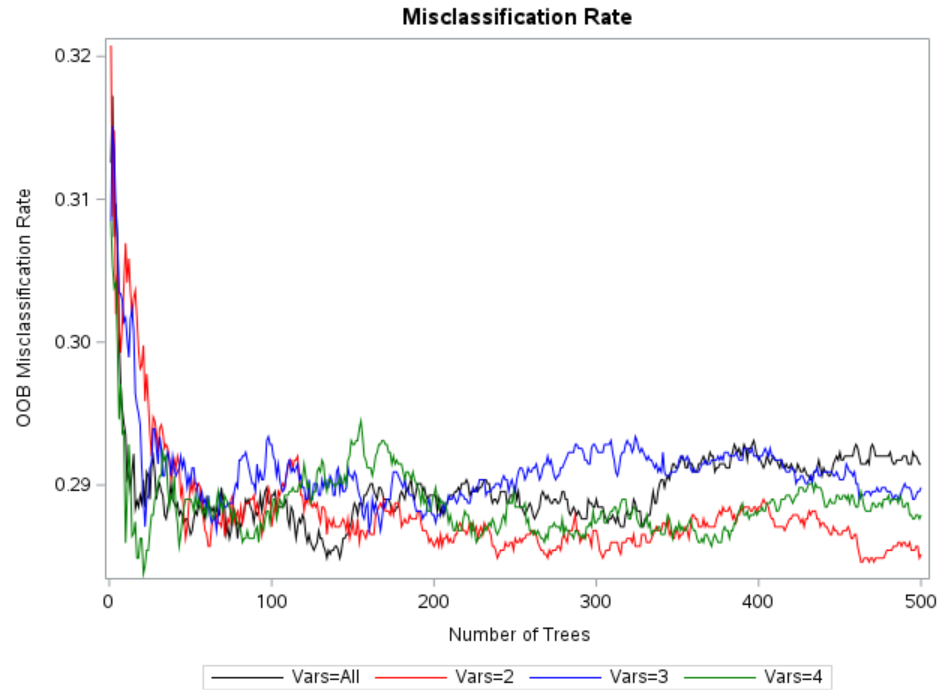
The following code tries different values in the VARS_TO_TRY= option and saves misclassification rate for each iteration. Later we figure out the optimal number of variables which have lowest misclassification rate.
%macro forest(mtry=);
proc hpforest data=heart_train maxtrees=500
vars_to_try=&mtry.;
target status/ level=binary;
input Weight Height AgeAtStart Smoking Sex / level = binary;
ods output FitStatistics = fitstats_vars&mtry.(rename=(Miscoob=VarsToTry&mtry.));
run;
%mend;
%forest(mtry=all);
%forest(mtry=2);
%forest(mtry=3);
%forest(mtry=4);
data fitstats;
merge
fitstats_varsall
fitstats_vars2
fitstats_vars3
fitstats_vars4;
label VarsToTryAll = "Vars=All";
label VarsToTry2 = "Vars=2";
label VarsToTry3 = "Vars=3";
label VarsToTry4 = "Vars=4";
run;
proc sgplot data=fitstats;
title "Misclassification Rate";
series x=Ntrees y = VarsToTryAll/lineattrs=(Color=black);
series x=Ntrees y=VarsToTry2/lineattrs=(Color=red);
series x=Ntrees y=VarsToTry3/lineattrs=(Color=blue);
series x=Ntrees y=VarsToTry4/lineattrs=(Color=green);
yaxis label='OOB Misclassification Rate';
run;
As per the graph above, VARS_TO_TRY=2 has lowest misclassification rate. Hence it should be used while building the model.


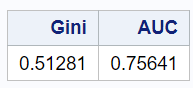

Share Share Tweet