This tutorial explains the various methods to split a dataset into training and test (or validation) sets in SAS.
Splitting a dataset into training and test sets is used for evaluating predictive models. The training dataset is used to build your model while the test dataset is used for assessing performance of the model on unseen data, checking for issues like overfitting.
Method 1 : Simple Random Sampling
By default, the PROC SURVEYSELECT procedure performs simple random sampling. The following code splits the dataset named 'heart': 70% of the data goes into the training dataset named 'heart_train' while the remaining 30% goes into the test dataset named 'heart_test'.
proc surveyselect data=sashelp.heart rate=0.7 outall out=heart2 seed=1234; run; data heart_train heart_test; set heart2; if selected =1 then output heart_train; else output heart_test; drop selected; run;
seed=1234: This sets the random number seed for reproducibility. It ensures that the same split is generated in the random sampling every time you run the code.
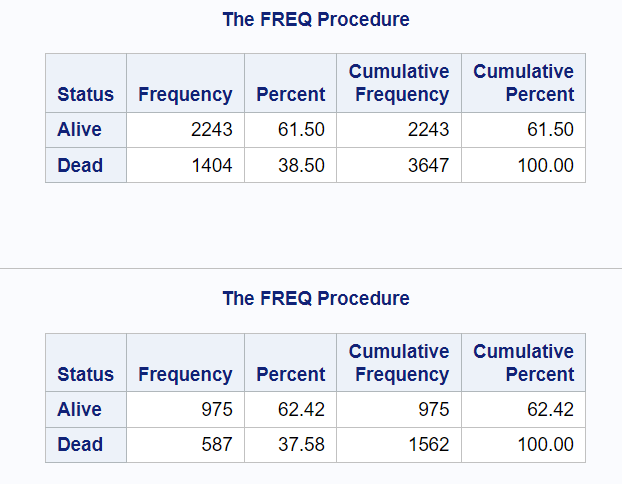
To validate the split, you can run PROC FREQ to see the number of observations in these two datasets along with the distribution of dependent variable.
proc freq data=heart_train; table status; run; proc freq data=heart_test; table status; run;
If you observe the distribution (percent) of dependent variable (status), you would find it is not consistent in the training and test datasets. It is 38.5% in training dataset and 37.58% in test dataset. The difference is not huge but when working with imbalanced datasets where one category is significantly more prevalent than others, simple random sampling might lead to biased model performance evaluation. Stratified sampling, which is explained in the next part of the article, helps deal with this problem.
Method 2 : Stratified Sampling
The benefit of the stratified sampling is that it ensures that the distribution of your dependent variable remains the same when splitting data into training and test datasets.
The following SAS code performs a stratified random sampling using the PROC SURVEYSELECT procedure and then splitting the selected data into training and test datasets.
proc sort data= sashelp.heart out=heart; by status; run; proc surveyselect data=heart rate=0.7 outall out=heart2 seed=1234; strata status; run; data heart_train heart_test; set heart2; if selected =1 then output heart_train; else output heart_test; drop selected; run; proc freq data=heart_train; table status; run; proc freq data=heart_test; table status; run;
This line strata status; indicates that the variable "status" is used as the stratification variable for sampling. This means that the sampling process will be performed within each unique value of "status". Make sure tha strata variable is sorted before running the PROC SURVEYSELECT procedure.
The distribution of status is same in both the training and test datasets. It is 38.22% in both the datasets.
Method 3 : Simple Random Sampling using ranuni() Function
The following code shows how to split a dataset into training and testing datasets using the ranuni function. This function is used to generate a random number for each observation, which is then used to shuffle the dataset before the split.
data heart2; set sashelp.heart; n=ranuni(1234); run; proc sort data=heart2; by n; run; data heart_train heart_test; set heart2 nobs=nobs; if _n_<=.7*nobs then output heart_train; else output heart_test; run;
The nobs option is used to obtain the total number of observations in the heart2 dataset. The _n_ variable represents the current observation number in the data step. The if _n_ <= 0.7 * nobs condition is used to determine whether an observation belongs to the training dataset (heart_train). If the condition is met, the observation is sent to 'heart_train' dataset; otherwise, it will go to 'heart_test' dataset.



Share Share Tweet